The predictable task that eats up valuable time in outbreaks
How can we avoid a monumental duplication of effort?
In the early stages of a major outbreak, you can guarantee that dozens – if not hundreds – of research groups will all perform the same predictable task in a similarly inefficient way. First, they’ll each go to a popular publication database (e.g. PubMed or Google Scholar) and look for common epidemiological parameters for the pathogen in question. And if it’s a new infection, they’ll look for a closely related one.
Perhaps they want to know the incubation period (i.e. delay from infection to symptoms), so they can explore quarantine options. Or the serial interval (i.e. delay from one case getting ill to the person they infect getting ill) so they can estimate the reproduction number. Or the delay from hospital admission to recovery so they can estimate potential bed requirements.
If it’s an infection that’s caused outbreaks before, maybe they’ll find some studies that have estimated these parameters. So they’ll download the relevant papers, then try to extract the values they need. But often studies won’t have reported exactly what they need. Maybe they want the distribution of the incubation period, so they can work out a reasonable upper limit for a quarantine period. But the papers in question have only reported the mean value, perhaps with a 95% CI for the overall distribution. Or there might be several studies with differing values, meaning they’ll either have to pick one or extract them all then work out how to combine them.
Next, with the values in hand, the researchers will input them – generally manually – into their analysis code. They’ll specify the value or distribution they need, then – finally – they’ll be able to run their code and address the question they set out to answer. Meanwhile, everyone else is doing the exact same thing.
Talk to any epidemiologist who works on outbreaks, and they’ll almost certainty have done the above. But it’s a huge duplication of effort, and one that creates lots of opportunities for errors to creep in at each step of the process.
Surely there’s a better way?
Last week, our team were part of a workshop at the WHO Hub for Pandemic and Epidemic Intelligence in Berlin to try and make some progress in this area. It’s part of an ongoing ‘epiparameter’ collaboration, bringing together different groups to improve the extraction, storage and reuse of common epidemic parameters.
One component of the work is the excellent systematic reviews that a team at Imperial College have been doing for the WHO R&D Blueprint priority pathogens. With their new R package, epireview, it’s increasingly possible to pull out parameters of interest much, much faster than ever before. So far, the dataset includes Lassa, Marburg and Ebola.
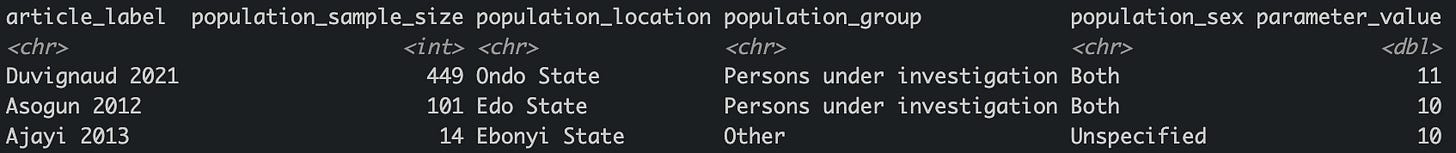
For example, here are studies reporting median duration of hospital stay for Lassa fever, using some quick filtering code:
The next step is using these parameters in an analysis. This is where the epiparameter tool comes in. This aims to make it easier to convert awkwardly reported values into usable objects for analysis. Rather than having to manually input distributions and values, we can increasingly build tools that use epiparameter functions, then let epiparameter and epireview do the hard work behind the scenes.
But what about infections that aren’t on the WHO priority pathogen list? My colleagues and I are gradually building out the database in epiparameter, but in the long-term it needs to be a much larger community effort, which the WHO Hub are co-ordinating.
This is all still a work-in-progress, but already tasks that would have taken days or weeks can now be done in minutes or hours instead. And once we have these databases of parameters and tools ready, there will be no need for future duplication of effort. Faced with a novel outbreak, teams can simply either use the existing resources, or contribute new estimates they’ve generated, so others can make use of them too.
There’s a lot of wider work happening in this area, from outbreak analysis to forecasting and scenario modelling. But all of these generally start with epidemiological parameters. So the hope is that in the early stages of a future major outbreak, you can guarantee that dozens – if not hundreds – of research groups will still perform the same predictable task. They’ll just do it in a much more efficient and collaborative way.


Adam, I am glad to read that we actually learnt and try to remedy what we have done wrong! I hope your team get the cooperation of the actors regardless of their political views. Thank you!
This is very encouraging. It's so easy to get really depressed about the state of public health, but this gives me some hope that there are some really important things going on that will make the next response better. Now if we can just get people to listen and trust us.